Difference between revisions of "Main Page"
(Created page with "Category:metabolite == Metabolite 2-METHYL-BUTYRYL-COA == * common-name: ** 2-methylbutanoyl-coa * smiles: ** ccc(c)c(=o)sccnc(=o)ccnc(=o)c(o)c(c)(c)cop(=o)(op(=o)(occ1(c(...") |
(Created page with "== SJAPGEM description == ''Saccharina japonica'' is the most cultivated alga worldwide, with nearly 8 millions tons of dry weight harvested in 2014, representing more than U...") |
||
| (6 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | == SJAPGEM description == | |
| − | == | + | |
| − | + | ''Saccharina japonica'' is the most cultivated alga worldwide, with nearly 8 millions tons of dry weight harvested in 2014, representing more than US $1.3 billion value ([http://www.fao.org/fishery/culturedspecies/Laminaria_japonica/en#tcNA008C Chen J., FAO website, accessed on january 10th, 2019]). | |
| − | + | Its draft genome sequence was published in 2015 ([http://doi.org/10.1038/ncomms7986 Ye et al., Nature Communications]). The genome version used here underwent an additional round of cleaning for bacterial contaminants using Taxoblast ([https://doi.org/10.7717/peerj.4073 Dittami and Corre, 2017]). The paper describing this GEM reconstruction in more detail was published in 2019 ([https://doi.org/10.3390/antiox8110564 Nègre et al., Antioxidants]). In this study, extensive manual curation was performed on the carotenoid biosynthesis pathway (CAROTENOID-PWY) and on the first cycle of xanthophylls (PWY-5945). In a follow-up study (Girard et al., in preparation) we are now curating the sterol biosynthesis pathways (PWY-8191, PWY-8238). | |
| − | + | ||
| − | + | == Automatic reconstruction with [https://aureme.genouest.org AuReMe] == | |
| − | * | + | Model summary: [[MEDIA:summary.txt|summary]] |
| − | ** | + | |
| − | * | + | Download '''AuReMe''' Input/Output [LINK OR MEDIA data] |
| − | ** | + | |
| − | + | The automatic reconstruction of ''Saccharina japonica'' results to a Genome scale [[MEDIA:S.japonica carotenoid curated.sbml|Model]] containing 3348 reactions, 3249 metabolites, 5018 genes and 1389 pathways. This GeM was obtained based on the following sources: | |
| − | + | * Based on annotation data: | |
| − | * [ | + | ** Tool: [http://bioinformatics.ai.sri.com/ptools/ Pathway tools] |
| − | * [ | + | *** Creation of a metabolic network containing 2551 reactions |
| − | * [[ | + | * Based on orthology data: |
| − | == | + | ** Tool: [http://pathtastic.gforge.inria.fr Pantograph] |
| − | * | + | *** From template ''NANNOCHLOROPSIS_SALINA'' ([https://doi.org/10.1186/s12918-017-0441-1 Loira et al., 2017]) creation of a metabolic network containing: 589 reactions |
| − | * [[ | + | *** From template ''ARABIDOPSIS_THALIANA'' ([https://doi.org/10.1104/pp.109.148817 De Oliveira dal'Molin et al., 2010]) creation of a metabolic network containing: 419 reactions |
| − | * | + | *** From template ''[https://gem-aureme.genouest.org/esilgem/ ECTOCARPUS_SILICULOSUS]'' creation of a metabolic network containing: 1492 reactions |
| − | * [[ | + | * Based on expertise: |
| − | + | ** 99 reaction(s) added | |
| − | + | * Based on gap-filling: | |
| − | + | ** Tool: [https://pypi.python.org/pypi/meneco meneco] | |
| − | + | *** 90 reaction(s) added | |
| + | |||
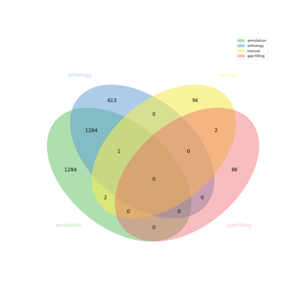
| + | [[FILE:venn.png|frameless|border]] | ||
| + | |||
| + | |||
| + | == Collaborative curation == | ||
| + | * Suggest reactions to add or remove: | ||
| + | ** Download this [[MEDIA:Add_delete_reaction.csv|form]] | ||
| + | * Suggest new reactions to create and add: | ||
| + | ** Download this [[MEDIA:Reaction_creator.csv|form]] | ||
| + | * '''Follow the examples given in the form(s) to correctly share your suggestions''' | ||
| + | * Send the filled form(s) to: gabriel.markov@sb-roscoff.fr | ||
Latest revision as of 11:18, 18 March 2021
SJAPGEM description
Saccharina japonica is the most cultivated alga worldwide, with nearly 8 millions tons of dry weight harvested in 2014, representing more than US $1.3 billion value (Chen J., FAO website, accessed on january 10th, 2019). Its draft genome sequence was published in 2015 (Ye et al., Nature Communications). The genome version used here underwent an additional round of cleaning for bacterial contaminants using Taxoblast (Dittami and Corre, 2017). The paper describing this GEM reconstruction in more detail was published in 2019 (Nègre et al., Antioxidants). In this study, extensive manual curation was performed on the carotenoid biosynthesis pathway (CAROTENOID-PWY) and on the first cycle of xanthophylls (PWY-5945). In a follow-up study (Girard et al., in preparation) we are now curating the sterol biosynthesis pathways (PWY-8191, PWY-8238).
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output [LINK OR MEDIA data]
The automatic reconstruction of Saccharina japonica results to a Genome scale Model containing 3348 reactions, 3249 metabolites, 5018 genes and 1389 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: Pathway tools
- Creation of a metabolic network containing 2551 reactions
- Tool: Pathway tools
- Based on orthology data:
- Tool: Pantograph
- From template NANNOCHLOROPSIS_SALINA (Loira et al., 2017) creation of a metabolic network containing: 589 reactions
- From template ARABIDOPSIS_THALIANA (De Oliveira dal'Molin et al., 2010) creation of a metabolic network containing: 419 reactions
- From template ECTOCARPUS_SILICULOSUS creation of a metabolic network containing: 1492 reactions
- Tool: Pantograph
- Based on expertise:
- 99 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 90 reaction(s) added
- Tool: meneco