Difference between revisions of "Main Page"
(Created page with "Category:reaction == Reaction RXN-15830 == * direction: ** left-to-right * common-name: ** cytochrome-c oxidase * ec-number: ** [http://enzyme.expasy.org/EC/1.9.3.1 ec-1.9...") |
(Created page with "== ESILGEM description == ''Ectocarpus siliculosus'' is a model filamentous brown alga. It was the first brown alga to be sequenced ([doi:10.1038/nature09016 Cock et al., Na...") |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | == ESILGEM description == | |
| − | == | + | |
| − | + | ''Ectocarpus siliculosus'' is a model filamentous brown alga. It was the first brown alga to be sequenced ([doi:10.1038/nature09016 Cock et al., Nature 2010]), and the genome sequence was used to build the first brown algal genome-scale metabolic network ([doi:10.1111/tpj.12627 Prigent et al., The Plant Journal, 2014]), using a preliminary version of the AuReMe pipeline. A second version of the genome-scale metabolic network, based on an improved version of the genome assembly and annotation ([https://doi.org/10.1111/nph.14321, Cormier et al., New Phytol 2017 ] was published in 2018 ([https://doi.org/10.1371/journal.pcbi.1006146 Aite et al., Plos Comp. Biol]). Now, in a more focussed follow-up study (Girard et al., ''in preparation''), we are curating specifically the sterol biosynthesis pathways (PWY-8191, PWY-8238). | |
| − | + | ||
| − | + | ||
| − | + | == Automatic reconstruction with [http://aureme.genouest.org AuReMe] == | |
| − | + | Model summary: [[MEDIA:summary.txt|summary]] | |
| − | + | ||
| − | + | Download '''AuReMe''' Input/Output [LINK OR MEDIA data] | |
| − | + | ||
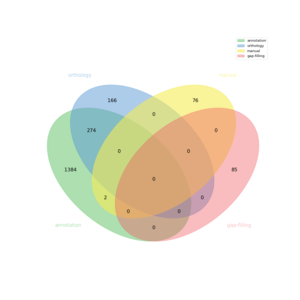
| − | + | The automatic reconstruction of ''Ectocarpus siliculosus'' results to a Genome scale [[MEDIA:model.xml|Model]] containing 1987 reactions, 2140 metabolites, 2288 genes and 1103 pathways. This GeM was obtained based on the following sources: | |
| − | * | + | * Based on annotation data: |
| − | ** | + | ** Tool: [http://bioinformatics.ai.sri.com/ptools/ PathwayTools] |
| − | *** | + | ** Creation of a metabolic network containing 1661 reactions |
| − | + | * Based on orthology data: | |
| − | ** | + | ** Tool: [http://pathtastic.gforge.inria.fr Pantograph] |
| − | *** | + | ** Creation of a global metabolic network containing 440 reactions |
| − | * | + | *** From template ''aragem'' creation of a metabolic network containing: 440 reactions |
| − | ** | + | * Based on gap-filling: |
| − | *** | + | ** Tool: [https://pypi.python.org/pypi/meneco Meneco] |
| − | + | *** 85 reaction(s) added | |
| − | + | * Based on expertise: | |
| − | + | ** 80 reaction(s) added | |
| − | == | + | |
| − | * | + | [[FILE:venn.png|frameless|border]] |
| − | + | ||
| − | + | ||
| − | + | == Collaborative curation == | |
| − | + | * Suggest reactions to add or remove: | |
| − | + | ** Download this [[MEDIA:Add_delete_reaction.csv|form]] | |
| − | + | * Suggest new reactions to create and add: | |
| − | + | ** Download this [[MEDIA:Reaction_creator.csv|form]] | |
| − | + | * '''Follow the examples given in the form(s) to correctly share your suggestions''' | |
| − | + | * Send the filled form(s) to: gem-aureme[at]inria.fr | |
| − | |||
Latest revision as of 09:01, 18 March 2021
ESILGEM description
Ectocarpus siliculosus is a model filamentous brown alga. It was the first brown alga to be sequenced ([doi:10.1038/nature09016 Cock et al., Nature 2010]), and the genome sequence was used to build the first brown algal genome-scale metabolic network ([doi:10.1111/tpj.12627 Prigent et al., The Plant Journal, 2014]), using a preliminary version of the AuReMe pipeline. A second version of the genome-scale metabolic network, based on an improved version of the genome assembly and annotation (Cormier et al., New Phytol 2017 was published in 2018 (Aite et al., Plos Comp. Biol). Now, in a more focussed follow-up study (Girard et al., in preparation), we are curating specifically the sterol biosynthesis pathways (PWY-8191, PWY-8238).
Automatic reconstruction with AuReMe
Model summary: summary
Download AuReMe Input/Output [LINK OR MEDIA data]
The automatic reconstruction of Ectocarpus siliculosus results to a Genome scale Model containing 1987 reactions, 2140 metabolites, 2288 genes and 1103 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: PathwayTools
- Creation of a metabolic network containing 1661 reactions
- Based on orthology data:
- Tool: Pantograph
- Creation of a global metabolic network containing 440 reactions
- From template aragem creation of a metabolic network containing: 440 reactions
- Based on gap-filling:
- Tool: Meneco
- 85 reaction(s) added
- Tool: Meneco
- Based on expertise:
- 80 reaction(s) added