Main Page
COKAGEM description
Cladosiphon okamuranus is an edible brown alga cultivated around Okinawa, with 20 000 tons of fresh weight harvested yearly, representing around 4 billion japanese yens in 2006. Its draft genome sequence was published in 2016 (Nishitsuji et al., DNA Research). The paper describing this GEM reconstruction in more detail was published in 2019 (Nègre et al., Antioxidants). In this study, extensive manual curation was performed on the carotenoid biosynthesis pathway (CAROTENOID-PWY) and on the first cycle of xanthophylls (PWY-5945). In a follow-up study (Girard et al., in preparation) we are now curating the sterol biosynthesis pathways, and also the biosynthesis of mozukulins, specialized steroids so far identified only in Cladosiphon okamuranus (also known as "mozuku" in Japan).
The photograph used here is cropped from a work by Mr. Kenji Iwa, published as CC BY SA 4.0. The original version can be accessed in this paper.
Automatic reconstruction with AuReMe
Model summary: summary
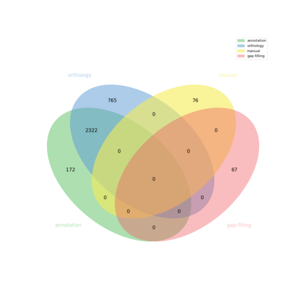
The automatic reconstruction of Cladosiphon okamuranus results in a Genome-scale Metabolic Model containing 3390 reactions, 2255 metabolites, 5373 genes and 1403 pathways. This GeM was obtained based on the following sources:
- Based on annotation data:
- Tool: Pathway tools
- Creation of a metabolic network containing 2494 reactions
- Tool: Pathway tools
- Based on orthology data:
- Tool: Pantograph
- From template ECTOCARPUS_SILICULOSUS creation of a metabolic network containing: 1468 reactions
- From template NANNOCHLOROPSIS_SALINA creation of a metabolic network containing: 595 reactions
- From template SACCHARINA_JAPONICA creation of a metabolic network containing: 3020 reactions
- From template ARABIDOPSIS_THALIANA creation of a metabolic network containing: 426 reactions
- Tool: Pantograph
- Based on expertise:
- 64 reaction(s) added
- Based on gap-filling:
- Tool: meneco
- 67 reaction(s) added
- Tool: meneco